We're a global volunteer team of students and scientists with a mission to help bring genomics to low-income countries.
Team accomplishments:
- First volunteer group to enable SARS-CoV-2 variant identification, demonstrating a low-cost solution for the global south
- First on-site sequencing of any organism in Mindanao, Philippines.
|
In collaboration with Philippine Genome Center Mindanao and the GECO-Philippines Project by the Research Institute of Tropical Medicine (RITM) and the University of Glasgow
|
|
Special thanks to our trainers (see videos here) Ineke Knot (portablegenomics.org); Dr. Nicole Wheeler; Dr. Amanda Warr; Dr. Joe Russell; Dr. Lara Urban; Ralf Dagdag and Matthew Redlinger from the Bortz Virology Lab; Alberten Lab; Dr. Nikki Freed; Dr. Michael Crone, and to our logistics consultants Jon Gotiong and Russel Ong.
Contact Kahlil Corazo via email [email protected] or Twitter DM @kcorazo if you want to collaborate or if you want to run a project to also bring genomic pathogen surveillance to a lab in your locality—we'll share with you what we've learned through project Accessible Genomics.
Contact Kahlil Corazo via email [email protected] or Twitter DM @kcorazo if you want to collaborate or if you want to run a project to also bring genomic pathogen surveillance to a lab in your locality—we'll share with you what we've learned through project Accessible Genomics.
|
Project introduction video
Project crowdfunding video (donate here)
|
Project Updates
Signup for project updates and training! Thank you!You have successfully joined our subscriber list. |
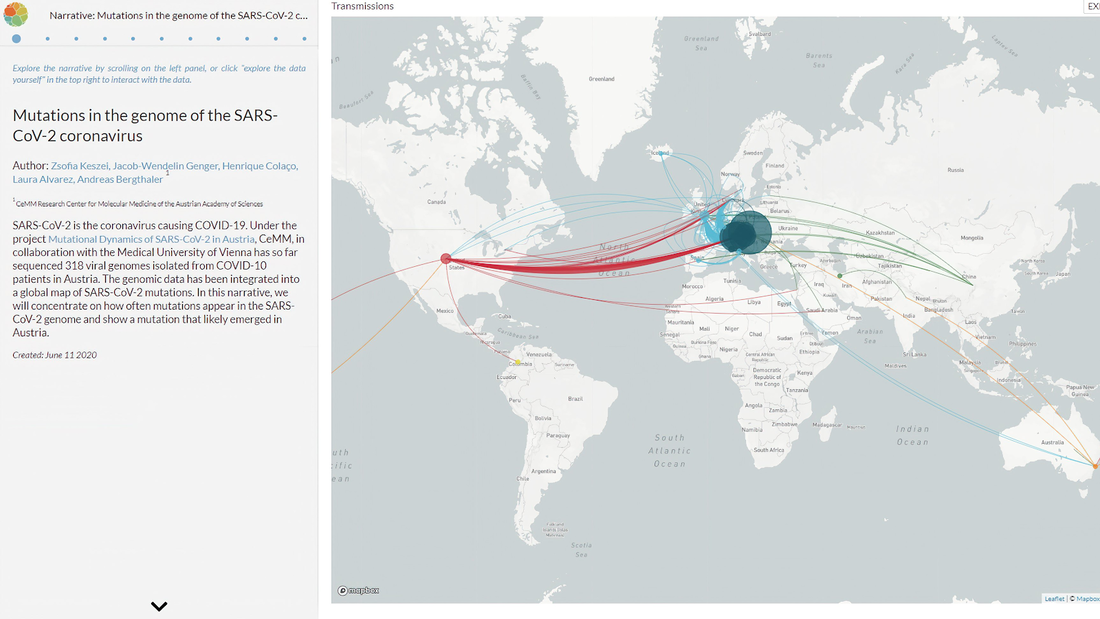
We need to bring genomic pathogen surveillance to the global south.To better manage this pandemic and prevent future ones, genomic pathogen surveillance is needed on top of diagnostics.[1]
RT-PCR and antibody tests could only tell us whether or not someone has an infection. Viral genomics could tell us the lineage of a specific infection. Tracing viral lineages is like 23andMe for pathogens. It would tell us where the virus originated. This information guides epidemiologist and local officials to make better decisions for quarantines and border management. The same skill set and equipment allows rapid identification of novel pathogens. |
image source: nextstrain
|
Access to genomics is currently limited to rich countries.
Yet, the next epidemics will mostly likely come from developing countries.
|
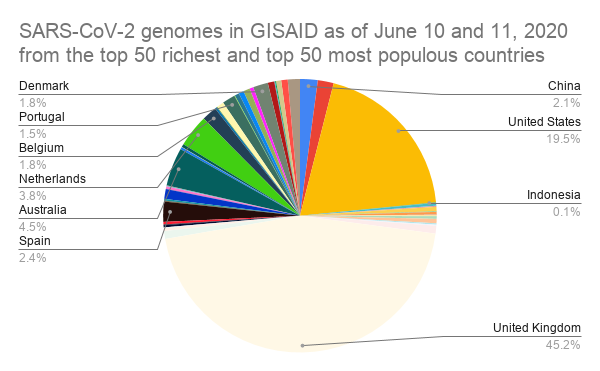
We counted the number of SARS-CoV-2 genome uploads per country in GISAID.[2]
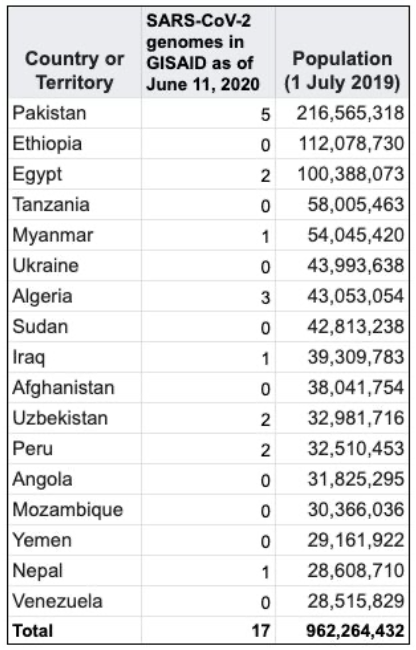
Almost half of the uploaded genomes came from the UK and a third of the rest were from the USA. At the bottom of this list are 17 countries with each less than 10 SARS-CoV-2 genomes uploaded in GISAID. The combined populations of these countries total to roughly 1 billion people. Future outbreaks will mostly likely come from the developing world, where human-to-wildlife interaction is most prevalent.[3] Yet, access to genomics concentrated in the developed world. |
Recent innovations allow us to bring genomics to the global south
|
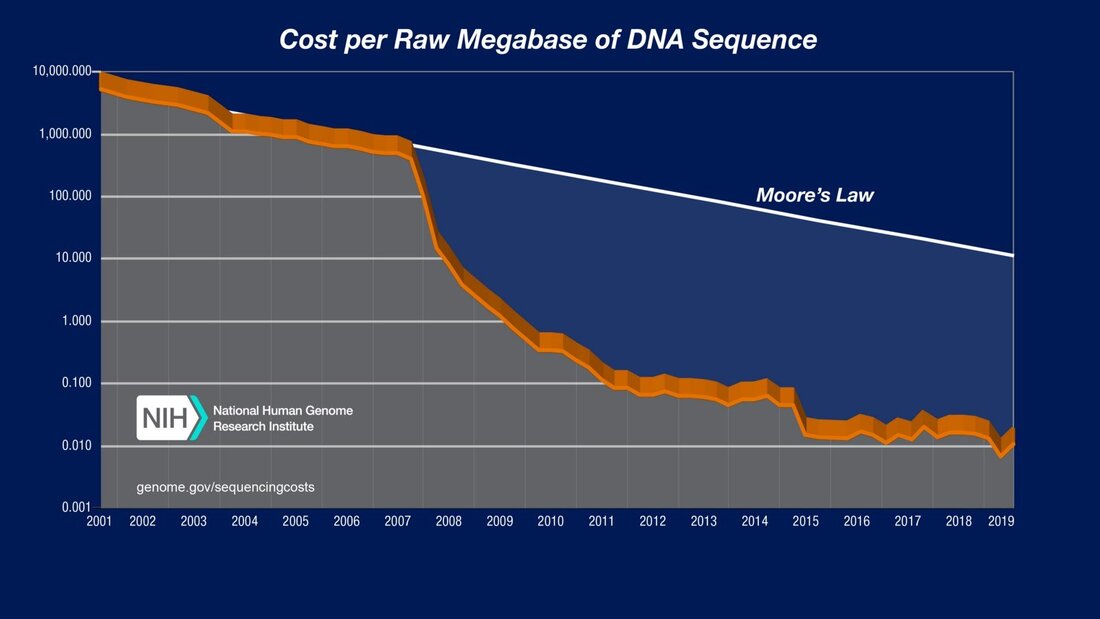
Just a few years ago, a genetic sequencing machine would cost more than a car, and a genomics lab hundreds of thousands of dollars.[4]
Today, there is a sequencer that costs less than an iPhone, the Oxford Nanopore MinION. The supporting lab equipment and consumables costs less than USD 20k.[5] After the lab is setup, the cost per sample is at $30 - $50.[6] Multiple scientists have shown the feasibility of using these new technologies in austere environments.[7] There are also well-established protocols, particularly for SARS-CoV-2.[8] This means that is is now possible for labs, universities and local governments in the developing world to setup a genomic surveillance laboratory. What is stopping developing countries then? Here are the challenges that each deployment needs to face:
|
Our contribution will be to create a deployment manual
|
Scientists and technologists will continue to lower the cost of sequencing and refining the protocols for its use.
To scale the deployment of these technologies, and to bring genomic pathogen surveillance to the global south, we also need project management solutions. A standard solution employed by institutions like Care.org and companies like Oracle is to establish a deployment manual. This manual documents tried-and-tested steps to manage a deployment, and could be used for training, and as a reference for project managers. Project ACCESSIBLE GENOMICS will produce this manual by deploying a genomic pathogen surveillance lab in a city in the developing world, and documenting the experience using the standards of the Project Management Body of Knowledge (PMBOK). |
We have assembled a team with the right science and project management backgrounds to successfully deliver this project
Project Accessible Genomics was born in the middle of the COVID-19 pandemic, at Just One Giant Lab (JOGL.io), a platform for open and collaborative international science projects.
|
Project Management
|
Subject Matter Experts
|
Accessible Genomics is a project led from the global south, for the global south, supported by a global team.
|
Go deeper
- Project Accessible Genomics Grant Application Template (details galore)
- Key papers for Project Accessible Genomics
- portablegenomics.org (portable genomics tools that could be used in a field setting, curated by Ineke Knot)
References
[1] To better manage this pandemic and prevent future ones, genomic pathogen surveillance is needed on top of diagnostics
[2] We counted the number of SARS-CoV-2 genome uploads per country in GISAID.
[3] Future outbreaks will mostly likely come from the developing world, where human-to-wildlife interaction is most prevalent.
[4] Just a few years ago, a genetic sequencing machine would cost more than a car, and a genomics lab hundreds of thousands of dollars.
[5] The supporting lab equipment and consumables costs less than USD 20k.
[6] After the lab is setup, the cost per sample is at $30 - $50.
[7] Multiple scientists have shown the feasibility of using these new technologies in austere environments.
[8] There are also well-established protocols, particularly for SARS-CoV-2.
- Examples of how developed countries use genomic pathogen surveillance in epidemic management
- “Studies already show that outbreaks tend to be shorter and smaller when genomics is used to help contact tracing.”
[2] We counted the number of SARS-CoV-2 genome uploads per country in GISAID.
[3] Future outbreaks will mostly likely come from the developing world, where human-to-wildlife interaction is most prevalent.
[4] Just a few years ago, a genetic sequencing machine would cost more than a car, and a genomics lab hundreds of thousands of dollars.
- Access to genomics might follow the history of mobile phone penetration. At the start, only developed countries had a significant percentage of their population with mobile phones. Now, the developing world has the most phones in the world.
- A sequencer used to cost more than a brand new car. Now, you can get the Oxford Nanopore MinION at less than the cost of an iPhone.
[5] The supporting lab equipment and consumables costs less than USD 20k.
- Bill of materials based on the ARTIC protocol
- Twitter thread on this bill of materials
- Section of grant application template discussing this bill of materials
[6] After the lab is setup, the cost per sample is at $30 - $50.
[7] Multiple scientists have shown the feasibility of using these new technologies in austere environments.
[8] There are also well-established protocols, particularly for SARS-CoV-2.
Signup for project updates and training!
Thank you!
You have successfully joined our subscriber list.