|
Hi folks! I have interviewed 6 scientists across 4 labs in 2 countries, Ghana and Cameroon:
When we started Project Accessible Genomics, our assumption was that the best way for us to help labs in the developing world establish SARS-CoV-2 sequencing operations is to pilot this in one lab and share our experience with them through a deployment playbook. Based on our experience with PGC Mindanao and my interviews with these labs in Africa, our problem assumption was incorrect. Even if we give labs a deployment playbook, it is probably unlikely that they can establish sequencing operations. The common barrier that I have heard from these labs in Cameroon and Ghana is that they don't have the capacity to get funding. They are running a lab, and writing grant applications is a high-effort, low-return endeavor. They still do as much as they can. Huguette, for instance, says that she only attempted to apply for 5 grants in the past year, and did not pass first screening of all 5. As we have experienced in our own grant applications for Project Accessible Genomics, each grant-giving body has its own language and bureaucracy that you need to learn to navigate. To frame the problem as capacity and capability to raise grant money is probably not helpful. It limits our solutions, and limits it to solutions that our team is not uniquely capable of delivering. I think the better framing is this: people in labs in the developing world just don't have slack. This is a better framing because it can allow different kinds of problems (ie, different kinds of lack of slack) and thus different solutions. The labs in Cameroon and Ghana lack money and time. PGC Mindanao turns out to have more access to money than the African labs I talked to (my guess is the same level as Kenya), but they also have a lack of slack in some other way. They have to work within the bureaucracy of the Philippine government, and inter-institution politics. So what happened in the past year in this project was really just a bunch of people with slack around the world directing that slack to a lab to accomplish a common goal we have. The scientists in our team had slack in terms of sequencing knowledge, JOGL was a channel of European financial slack, and each team member had some time and skill we chose to channel for the project. By the way, here's a list of different forms of slack. This is more than just about money. (It is a good personal exercise to list for each category your source of slack and of tightness and plan out how to increase slack and reduce tightness.)
So the solution is not some knowledge base (or not just it). It is to find slack to help these labs. Now, where can we find this slack? And what could these people get in giving some of their slack? (Doing something good is a good reason, but you can't rely on that to sustain something.) I'm currently part of this iGEM team called Friendzymes. I'm helping out mainly by training them in project management. But I can't keep up with these kids. The Discord server is moving so fast. It made me realize that the people with the most slack are those who are still in school. They are not yet burdened by responsibilities of adulthood. Their parents essentially work so they can have slack, intended for their education. So here's my proposal. What if we form student teams for each of these labs? They figure out what kinds of problems need to be solved for each lab to enable them to sequence SARS-CoV-2. Through this experience, they get to make an impact and they get hands-on learning. This is essentially copying the model of iGEM but instead of genetic engineering, the students get to learn professional project management, and entrepreneurship skills. They'll also learn about genomics. Post-pandemic, this could be a program to solve other problems in lab in the global south, not just lack of genomic SARS-CoV-2 surveillance. The easiest way to do this is to plug in the program in iGEM or some other organization that has an existing base of high-slack individual who want to learn. Worst case, we can run the program ourselves. I've been doing project management training for the past 10 years. A lot of them were for student leaders. So I think I can do this solo if needed. Would love to hear your thoughts. Also, if you know anyone in iGEM or a similar organization we could talk to about doing this kind of program, I'd appreciate an introduction.
0 Comments
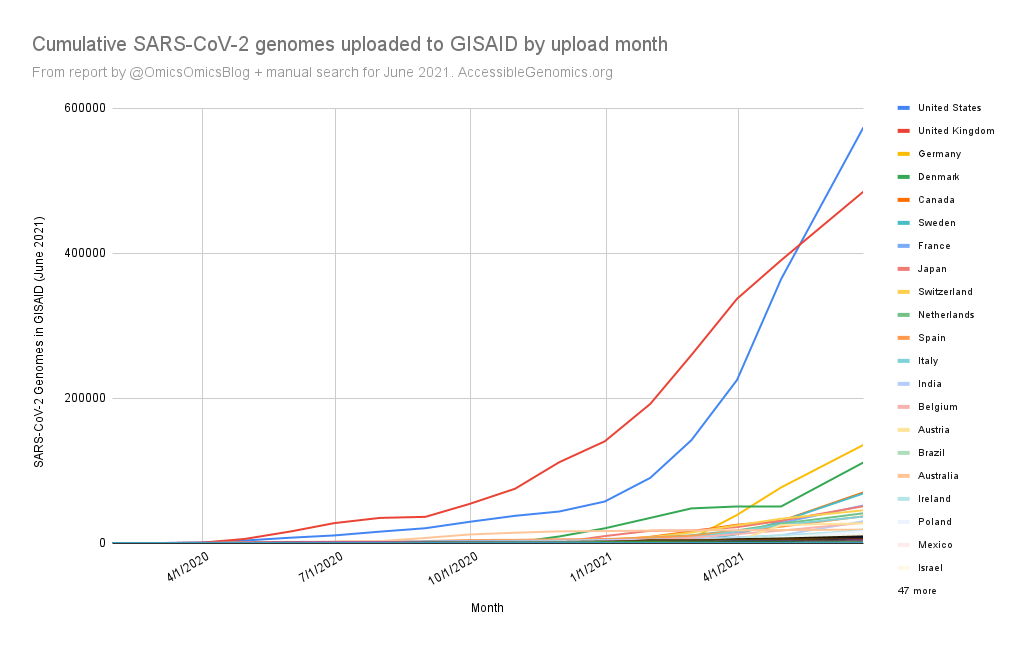
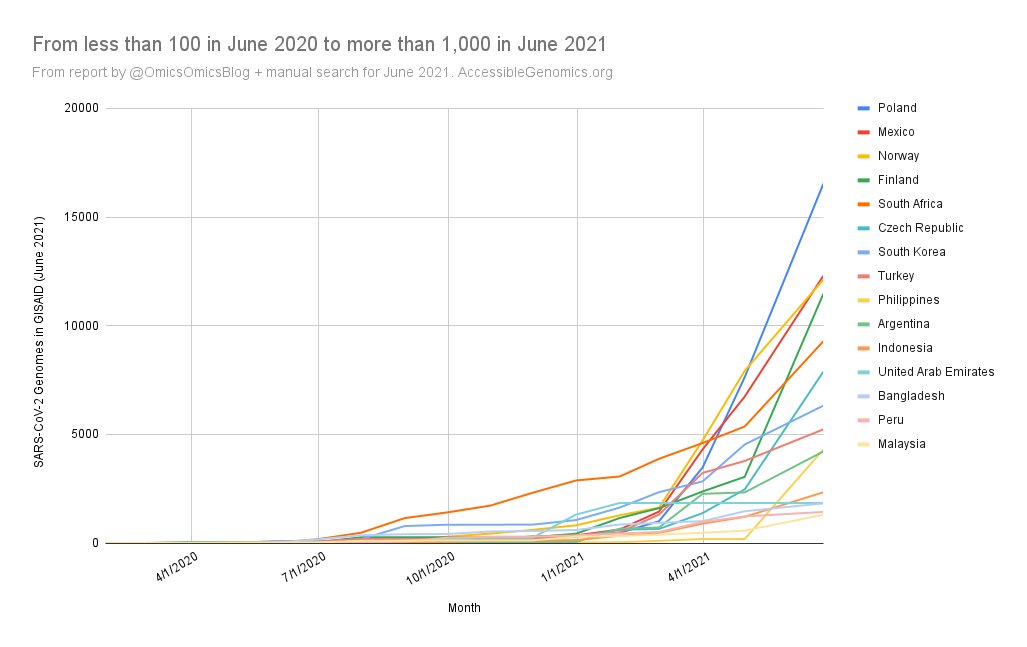
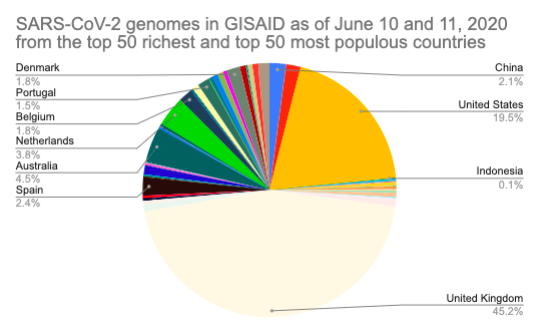
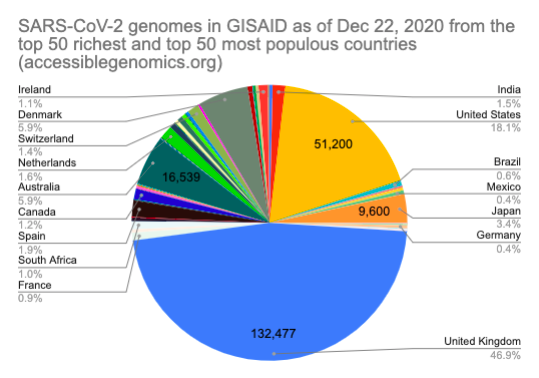
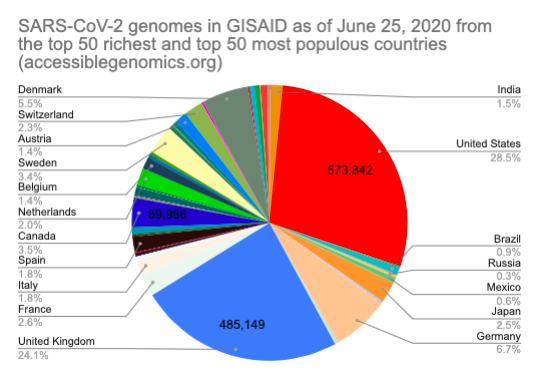
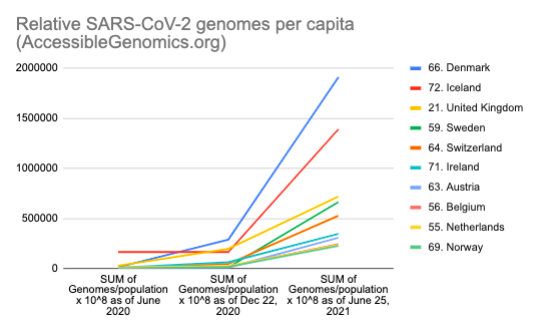
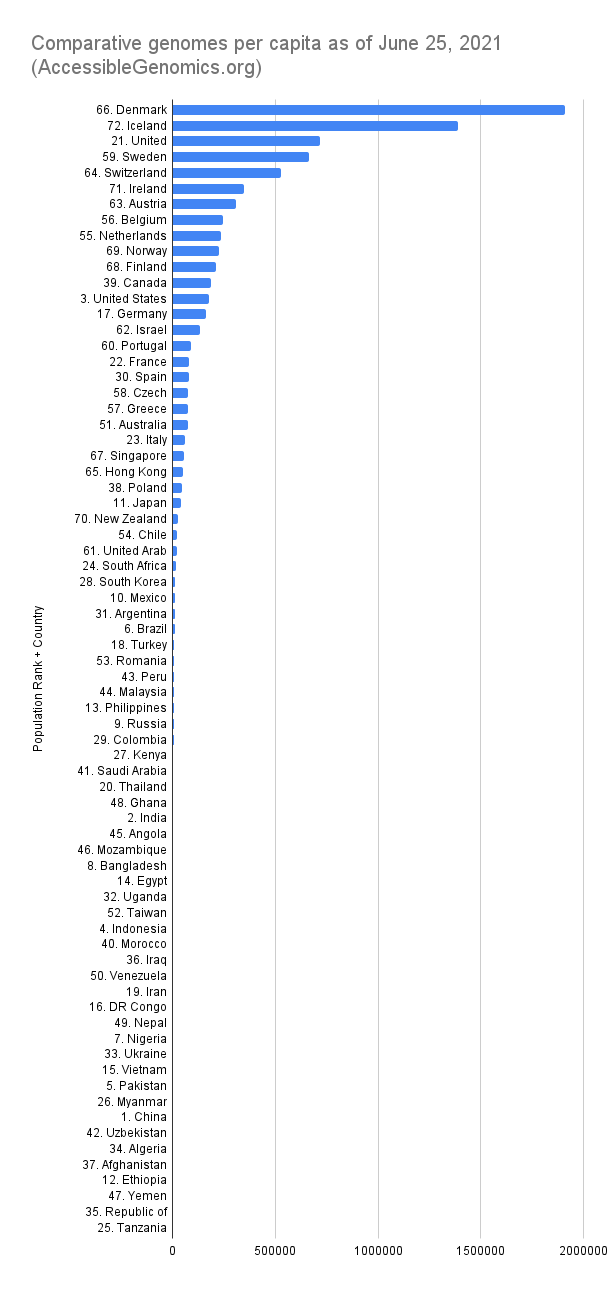
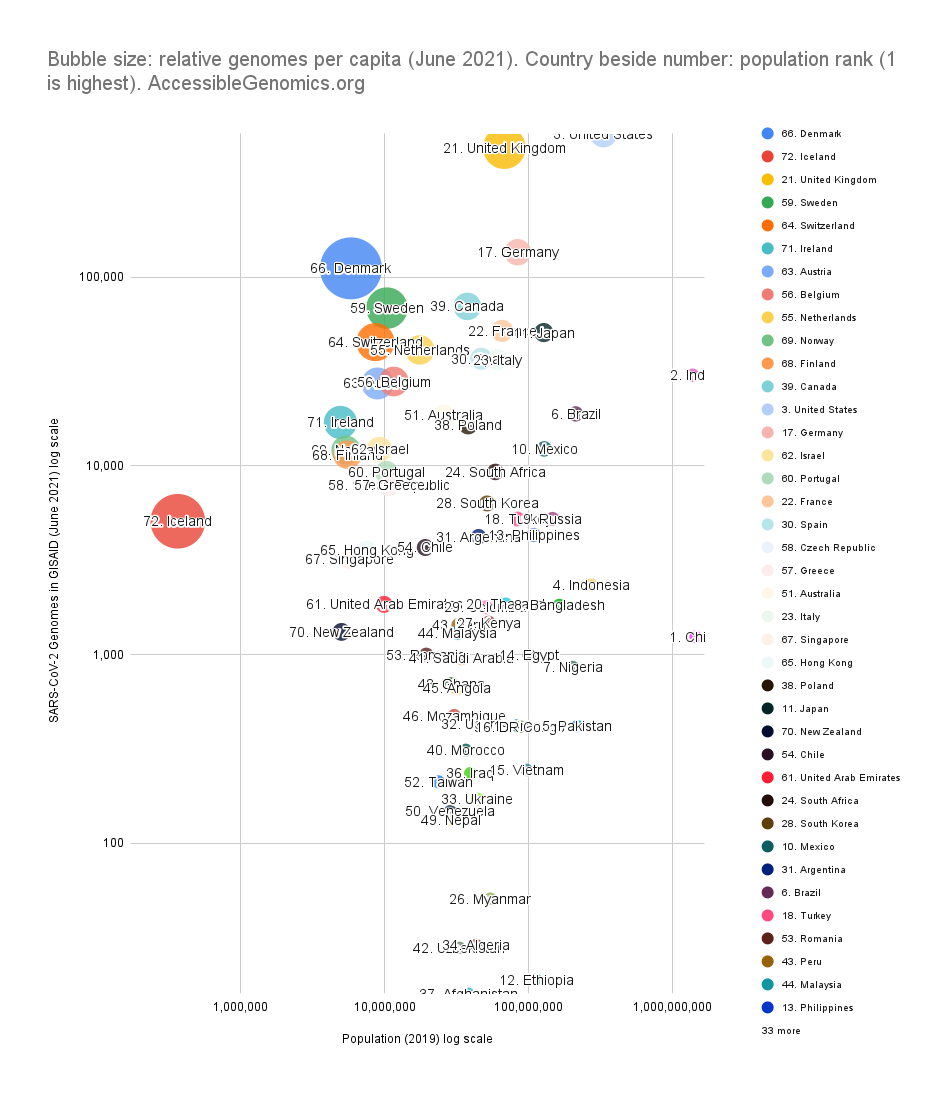
By Kahlil Corazo HighlightsGISAID.org just reached 2 Million SARS-CoV-2 genomes. More than a year after the pandemic started, the USA has overtaken the UK in terms of the number of SARS-CoV-2 genomes shared. The US and the UK combined still represent more than 50% of the shared genomes. However, eight countries still have less than 100 genomes in GISAID. Three have 0. Denmark still leads in terms of genomes per capita, followed by Iceland, then the UK. For Denmark and Iceland, it appears that it only took one lab to make a difference. Some countries have made huge strides. In June 2020, there were 38 countries with less than 100 genomes shared. Now, there are only eight. My country, the Philippines, went from 17 SARS-CoV-2 genomes to 4,307 🎉. BackgroundThis is the third time I've reported SARS-CoV-2 upload numbers in GISAID. The first two were Twitter threads:
Here are my assumptions:
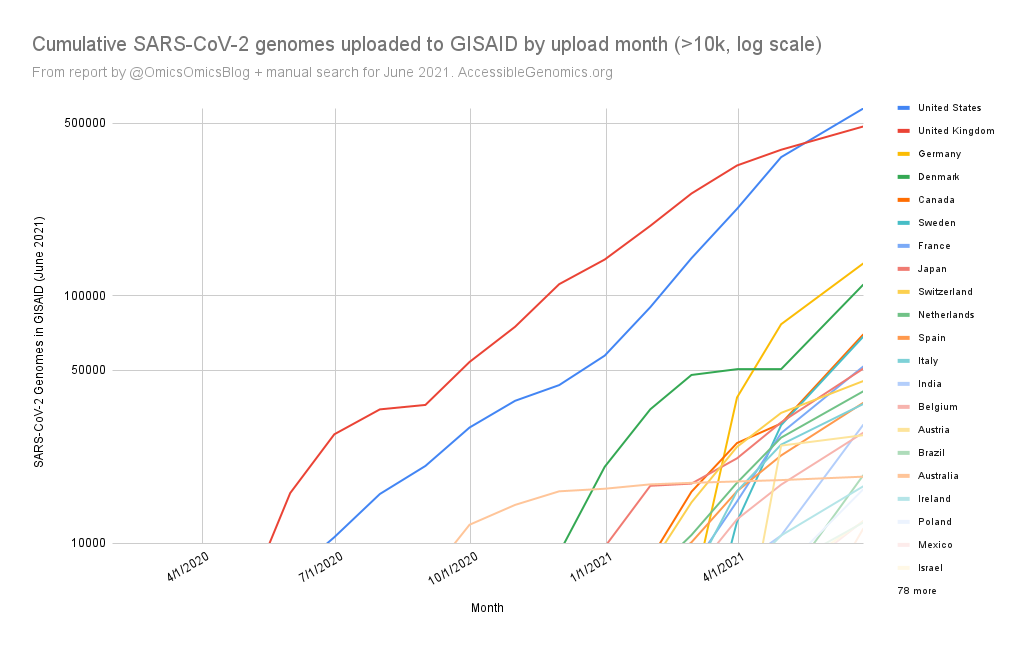
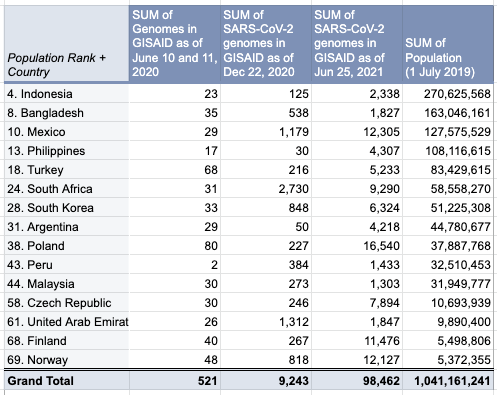
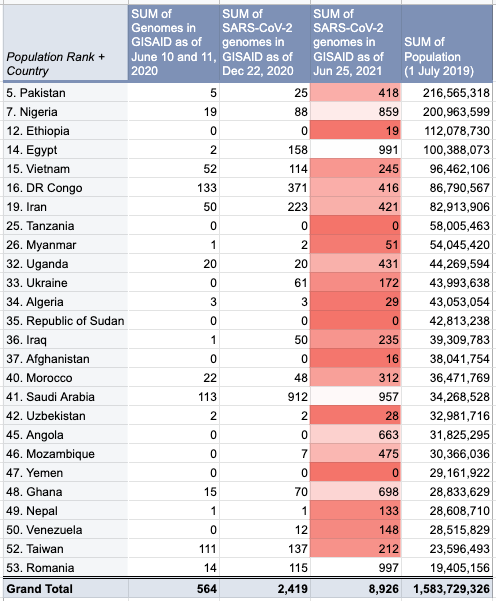
The report gave us an idea on which countries we should focus on for Project Accessible Genomics. Here's the spreadsheet I used. I manually reported the numbers (using country search). I don't have the chops, the bandwidth and the computing power to create reports straight from the database. I only included the 50 countries with the highest population plus the 50 countries with the highest GDP. Some of the graphs below are from a report created by Keith Robison of OmicsOmicsBlog, which has numbers up to March 2021. Here's the spreadsheet I used for that. If you are viewing those spreadsheets, apologies in advance for how messy they are. For a background on the importance of sequencing and sharing data to fight this pandemic and future outbreaks, here's a nice article by Time.com: What We Learned About Genetic Sequencing During COVID-19 Could Revolutionize Public Health. Okay, let's make some graphs! Absolute countsMost improved countries26 countries with less than 1,000 genomes sharedGenomes per capitaTeam accessible genomics got to do a Q&A with Albertsen Lab, which is the reason why Denmark has the highest genomes per capita. Here's the recording: www.youtube.com/watch?v=HTV9BIxmFtk
In Iceland's case, it is probably also because of one lab (I have not confirmed this). While Albertesen Lab is an academic lab, Iceland's deCODE, a commercial genomics company (a subsidiary of Amgen). The UK's leadership is probably due to its heavy investment in genomics in the 2000s' see twitter.com/kcorazo/status/1271010623544004608 |
Project Accessible GenomicsWe're a global volunteer team with a mission to bring genomics to low-income countries. ArchivesCategories |
 RSS Feed
RSS Feed